
|
Kirsten ten Tusscher |
| Home | Research | Publications | Source codes | Projects for Internships | |
Research
The research of the Ten Tusscher group focuses on the deciphering of
developmental patterning processes in both plants and animals. For this
the group uses state-of-the-art multi-scale modeling approaches and closely
collaborates with experimental research labs.
Our current research encompasses three major research lines:
- Plant root development and adaptation to environmental conditions
- Regulatory networks controlling cell fate decisions
- Development and evolution of animal body axis segmentation
Job opportunities
Computational Phd postion on modeling Arabidopsis resilience to drought and temperature stress
Crop-XR is a large Netherlands based initiative aimed at increasing sustainability of agricultural practices by making crops less dependent on fertilizers and pesticides and more resilient to climate change and environmental stress. For this an increased understanding of plant stress responses is essential. For this project we are looking for an enthusiastic PhD candidate to develop dynamical, process-driven computational models for the effects of combined temperature and drought stress on plant growth and performance. The successful candidate will develop state of the art functional-structural plant models simulating plant growth dynamics and integrate these with the molecular signalling networks linking drought and temperature perception to responses in plant physiology, growth and development. To arrive at the relevant molecular signalling networks the PhD candidate will collaborate with experimentalists and machine learning experts. Ultimate goal is to identify the interplay between temperature and drought stresses and pinpoint the molecular hubs at which trade-offs and synergies arise that can be modulated through targeted breeding approaches.
For this positions we are looking for candidates with an Msc degree in Computational Biology, Biophysics, Applied Mathematics or a related field that have a strong interest in plant science and are keen to bridge biological and computational approaches. For the project experience with differential equation based modeling and programming are a requisite, while affinity with data analysis and bioinformatics will be considered a bonus.
Plant root development
Unlike animals, plants keep growing and generating new organs throughout
their life span, with formation of each new organ involving the de novo
formation of a stem cell niche driving organ growth. Major research questions
involve which processes pattern the stem cell niche and meristimatic region
of dividing cells emanating from it, and which processes prepattern the
locations along the main root competent for the future formation of lateral
roots. In our research we use multi-scale cell-based models
incorporating gene expression, hormonal signalling as well as growth,
division expansion and differentiation of cells to answer these questions.
Using this approach we previously demonstrated a division of labor between
auxin and the downstream PLETHORA transcription factors in determining
meristem size and rates of division, elongation and expansion (Mahonen et al.,
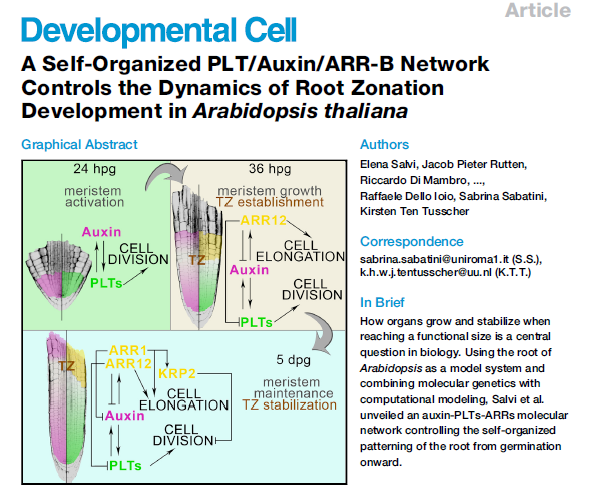
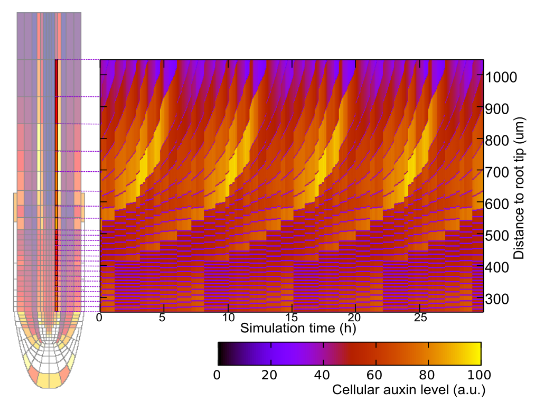
Nature, 2014). We subsequently recently demonstrated that the
auxin-PLETHORA-ARR network controls activation, outgrowth and stabilisation
of the root meristem after germination (Salvi et al, Dev Cell, 2020).
Additionally, we have shown how lateral root priming emerges from the synergy
between plant root tip auxin transport and root growth dynamics (van den Berg
et al, BioRxiv, 2018).

Plant development also differs from animal development in that it is
highly plastic, leading to non-stereotypical, environmentally dependent
plant architectures. As an example, plant roots show directional growth,
called tropisms, towards gravity but also away from salt. Additionally,
in response to heterogeneous nutrient supplies, plant root systems
show a preferential proliferation of roots in nutrient rich patches,
called preferential root foraging. Tropisms can still be effectively
studied by cell-based models of a single root, enabling us to succesfully
identify additional genes contributing to the auxin asymmetry underlying
root salt avoidance (van den Berg, Development,2016). However, to adress
which processes give rise to an asymmetric growth of different parts of
the root system in response to differences in their local nutrient
conditions we need to expand to spatially more coarse-grainded FSP-type
multi-scale models of overall root architecture. Using simplified
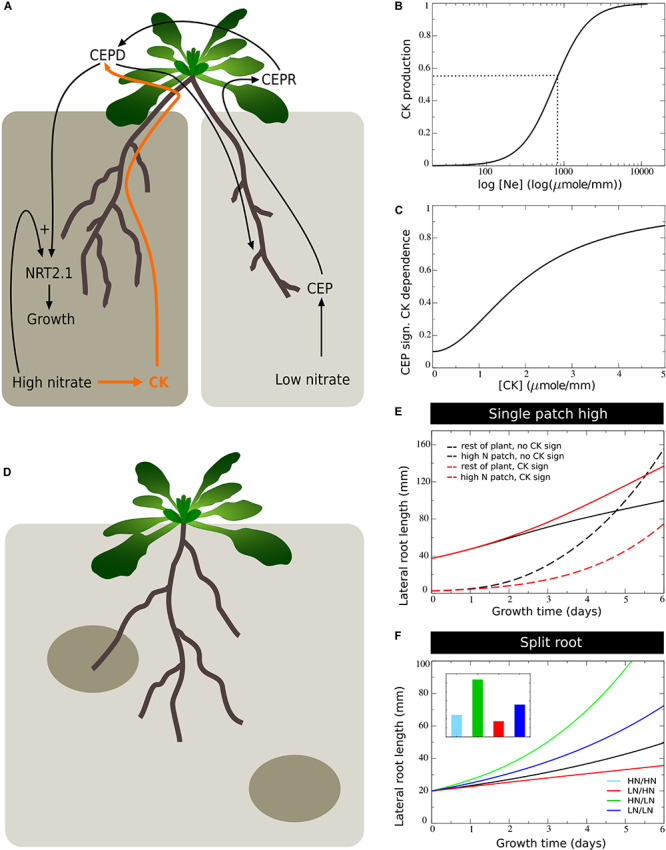
models of this type we have already demonstrated that local, long-range
and systemic nutrient signalling are likely insufficient to explain
preferential nitrate foraging, and that the competition between roots
for carbon resources further amplifies nutrient-difference induced
asymmetries (Boer et al., 2020). To further enhance the realism and
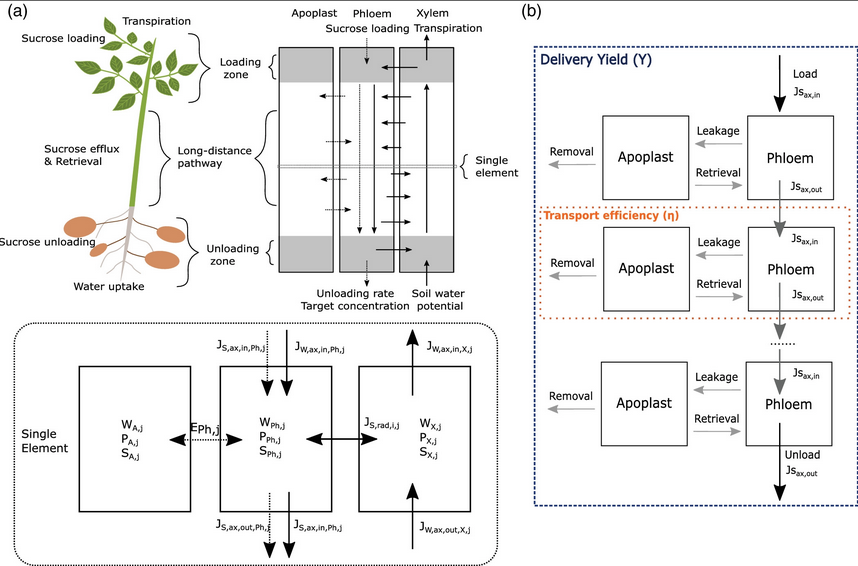
power of this overal root architecture modeling approach we recently
developed our own biophysical model for water and carbon transport
(van den Herik et al., Plant Cell Env, 2020) that can easily be integrated
in this framework.
Cell fate decision making
During development cells have to decide whether to keep dividing and
maintain an undifferentiated state, or rather differentiate and stop
dividing. Additionally, upon differentiation choices between alternative
cell fates have to be made. Importantly, in healthy non-cancer cells,
the decision to differentiate is irreversible and terminal differentiation
is mutually exclusive with an active division status. A key question is
how the architecture and dynamics of the regulatory networks -genetic,
epigenetic and posttranscriptional- controlling cell behavior give rise
to these decisions in cell fate.
To adress these questions, we recently started a project in collaboration
with the C.elegans groups of Sander van den Heuvel and Rik Korswagen,
as well as with the labs of Alexander van Oudenaarden en Michiel Vermeulen.
The overarching idea is to use single cell transcriptomics as well
as epigenetic and protein data to reverse engineer the architecture
and dynamic functioning of the network underlying mesoblast and neuron
differentiation in C. elegans.
By combining omics based network inference and differential equation
based modeling of gene expression dynamics, we aim to determine through
sophisticated fitting and optimization procedures the architecture
of the core regulatory networks involved.
Animal body axis segmentation
A segmented body axis occurs in three major animal clades, vertebrates, annelids and arthropods. In all three cases, the segments are prepatterned through an
oscillatory processes occuring in the posterior part of the body. Through growth these oscillations are transformed into a spatially periodic stripe-like pattern, giving rise to an anterior-to-posterior sequence of formed segments.
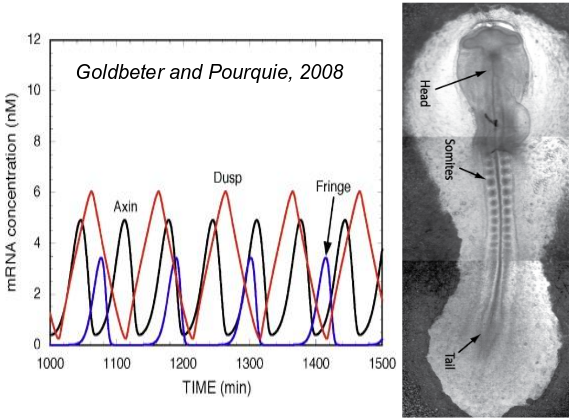
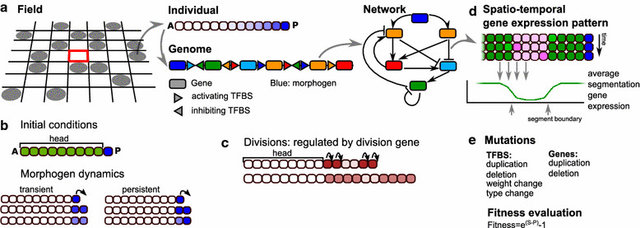
A major research question is whether this sequential segmentation mode was present in the urbilaterian ancestor and was subsequently lost in many animal clades or rather evolved independently in the clades. Additionally, it is unclear what the sequence of evolutionary innovations was that likely led to the innovation of segmented body plans.
To adress these questions, we use an in silico evo-devo approach, simulating the evolution of developmental processes in populations of simplified, multicellular developing organisms, each endowed with a genome encoding for a regulatory network controlling cell state. By replaying the in silico tape, varying mutation rates, selection pressures, and initial conditions, we investigate under which conditions segmentation evolves, the type of segmentation process evolving, and the order of evolutionary and mutational events.
Group members
Current members
- Kirsten ten Tusscher, Full professor, Principal Investigator
- Lucila Salvatore, PhD student, experiments on root nutrient decision making
- Eva van Zelm, postdoc, experiments on lateral root development in response to nutrients
- Bas van den Herik, postdoc, phenotyping and modeling of plant development and decisio making
- Jeroen Saccheri, PhD student, cell-based modeling of main and lateral root development
- Jerry Chen, PhD student, interplay plant physiology, ecology and climate
- Oscar Jordan, master student, deciphering the interplay of root halotropism and gravitropism
- Wouter van Etteger, master student, modeling C/N balance and decision making
Former members
- Thea van den Berg, PhD student, salt stress, lateral root priming
- Jaap Rutten, PhD student, role of auxin cytokinin interactions in root growth
- Milton Noguira da Silva Junior, PhD student, biophysics pf lateral root formation
- Xiang Zhang, postdoctoral researcher, mechanics of root growth
- Alexandros Skourtis Cabrera, master student, interplay of PXY-CLE in determining secondary root growth
- Jorian Flik, master student, deciphering the mechanism of root phototropims
- Joana Teixeira Santos, PhD student, effect of shade avoidance on root architecture
- Daniel Weisse, postdoctoral researcher, mechanics of root growth
- Sophia Scheper, Master student, evolution of segmentation
- Meine Boer, Master student, models of overall root system architecture
- Thijs Geurts, Master student, directional responsesin roots, halotropism
- Renske Vroomans, PhD student, evolution of segmentation in bilaterian animals
- Peter de Greef, Master student, phosphate starvation
- Bram van Dijk, Master student, Evolution of evolvability, genome versus network structuring
- Rutger Bos, Master student, Pro's and cons of thermodynamic models for evolutionary simulations
- Ioannis Tamvakis, Master student, Self-organised auxin patterning in plant development
- Klaartje van Berkel, PhD student, Self-organization of auxin-PIN patterning in plant development
- Jelmer de Ronde, Master student, Characterisation of the human ENCODE gene regulatory network
- Redmar van den Berg, Master student, Polymorphism and speciation in diploid genome-network models
- Janita Terpstra, Master student, Sympatric speciation in absence of assortative mating signals
- Harold Wolff, Master student, Reconciling models for planar cell polarity


